Eighty to ninety percent of a data science project involves processing the data, this has to happen before any model is fit. Data comes in a variety of disarray, different file formats, empty values, even no formatting. Once you sift through the data, organize it, standardize it, and really start to visualize the data, feature selection can begin.
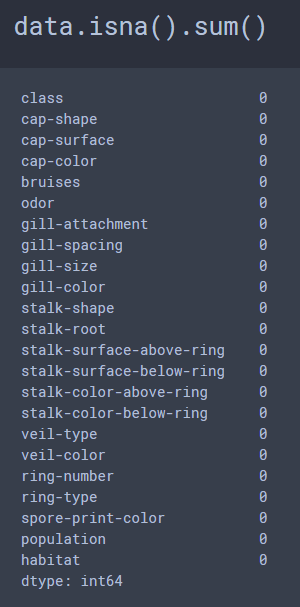
I am going to use the Mushroom Classification Dataset, which provides 8,124 samples, with 23 features for each sample. Unusually, there are no missing values for any of the features for any of the samples in this dataset and the dataset was very balanced, which I rarely encounter.
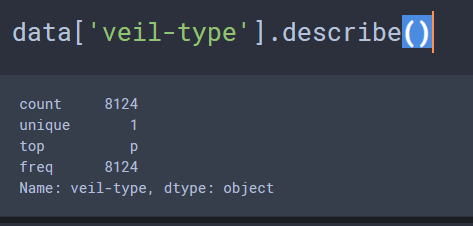
There are a variety of attributes being assessed for the binary classification task of whether a mushroom is poisonous or edible based on these 23 physical characteristics. Actually, lets make that 22 right off the bat, due to the fact that there are two possible values for the ‘veil-type’ attribute, partial=p & universal=u, however there is only 1 unique value for this column, so this column can be eliminated off the bat, due to the fact that 100% of the samples present in this data display the ‘partial’ veil-type attribute.
That was the easy part. I decided to try VERY basic feature selection based on the variance and also for correlation, to compare the features selected using each method. Now, since the data is categorical by nature, calculating the variance involves first label encoding the data.
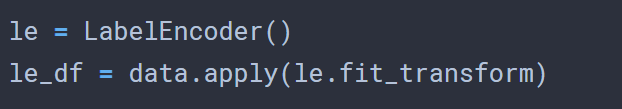
With a quick check on the variance for each column, simply within the column, I chose the 5 columns with the highest variability.
['stalk-color-below-ring', 'odor', 'spore-print-color', 'cap-color', 'gill-color']
When checking the correlation coefficients for each column based on the feature and the target, the following columns returned the highest correlation coefficients.
['stalk-root', 'ring-type', 'bruises', 'gill-color', 'gill-size']
When I noticed that ‘bruises’ was present in the second set of features, I had some questions, due to the fact that I am not an expert in mycelium, fungus or mushrooms, whatsoever. I found out that the presence of bruises isn’t what is critical in identification, but the color which the mushroom bruises. The dataset only provides the following regarding bruises: bruises=t, no=f, so it’s simply the presence, not the color. For now, I will leave this alone, however, I will be researching this further.
I ran GridSearchCV with a 5-fold StratifiedKFold() for cross validation for both sets of features.
The following are the results for the dataset with feature selection performed using correlation coefficients.
best parameters are: RandomForestClassifier(max_depth=8, n_jobs=-1)
best parameters are: KNeighborsClassifier(algorithm='ball_tree', weights='distance')
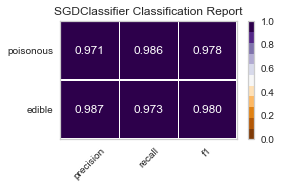
best parameters are: SGDClassifier()
best parameters are: SVC(C=1, degree=1)
Now these are the GridSearchCV results, with the same cross-validation on the variance selected features.
best parameters are: RandomForestClassifier(max_depth=8, n_jobs=-1)


best parameters are: KNeighborsClassifier(algorithm='ball_tree', weights='distance')
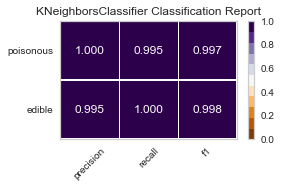
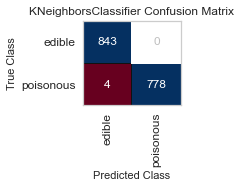
best parameters are: SGDClassifier()


best parameters are: SVC(C=1, degree=1)


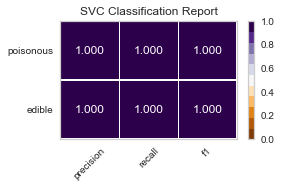
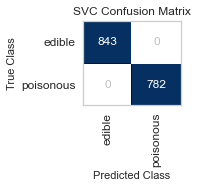
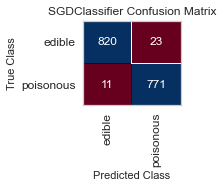
For both sets of feature selected datasets, I keep running into Type II, False Negative errors, which in this case are detrimental to the person eating the poisonous mushroom, thinking it to be edible. I will be further honing my feature selection, and doing more research to conclude the set of features that minimizes the Type II error for this classification task.
When I ran the same GridSearchCV on my label encoded data, my results for RandomForestClassifier() were far better, as no one was ill advised to eat a poisonous mushroom using all of the features(minus the veil-type feature I removed early on). Perhaps I need to reassess the limit of 5 features to classify the mushrooms edibility, and expand to 6 or 7, as that could be the one missing element.
best parameters are: RandomForestClassifier(max_depth=8, n_jobs=-1)


best parameters are: KNeighborsClassifier(algorithm='ball_tree', weights='distance')
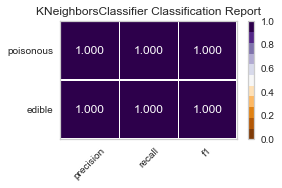

best parameters are: SGDClassifier()
best parameters are: SVC(C=1, degree=1)